IS Analyzer Project
An applied statistics project involving heaving webscraping and NLP processing
Hello Statistics!
This project spawned initially from an opportunity presented to me through my penultimate semester coursework. At the College of Wooster, I took DATA-230: Applied Statistics as a foray into the world of statistics and data science. 4-year-old high school statistics was really pulling its weight for me at the beginning of this course… ‘Applied Statistics’ is a course that has been just that – applied, primarily covering linear and logistic regressions in the context of applying and fitting various models to real world datasets. A substantial portion of the course has been building towards a free-form final project that is basically “go do statistics”. Of course, the project specification is sleighted towards working with existing datasets and building models on that pre-compiled data. As a computer scientist (and wanting an excuse to learn some webscraping and data processing), I instead immediately turned into the figurative brush in trying to first build my own dataset before using that data to play around with. In a sense, this meant I did non-trivial work just to get to the starting point of my other peers, but thanks to an ammenable instructor, we modified the expectations of the project enough such that I still did statistics while accounting for the extra effort done in step 0.
Pre-preamble… complete!
Motivation and Context
At the College of Wooster (my undergraduate institution), there is substantial importance placed on our “Independent Study” project. Our Independent Studies are year-long independent, individual research projects in the realm of each student’s chosen major(s). As a senior in the computer science department, I am working on my own IS – separate from this statistics project. Refer to other posts and pages on my site if you are interested in static analysis and understanding energy usage of programs. Back to an overview of ISes, each department differs somewhat, but generally each week seniors check in with an advisor in their department, who may or may not be an expert in the specific topic of the IS. To be pompous, Independent Study is a very souped up version of a senior thesis at other institutions. Although I do not believe college rankings and special ranklists are anywhere near honest, for the last 20-ish years, Wooster has been a premier undergraduate mentored research institution.
I am not experienced in the machine learning space, but with increasingly ease of accessibility of generative AI, for the last year or so I was playing around with a dream to tune a GPT to independent studies and see the results – for humor’s sake.
At this time, I was unaware of where I could collect IS training data, but at the beginning of the year I came across our college’s OpenWorks page.
Now I have a source for where to collect data, as there is an “Independent Studies” publications page on Openworks. I also was reviewing final project writeups at this time, and saw that my statistics class was very open ended. This is a perfect excuse to harvest all this data for personal project use for a course project. I just need to do the collection.
Webscraping, Go, Colly, and You
My day job involves a lot of Selenium Automation in Groovy. My first thought for webscraping is exactly that – lets do some kind of Selenium based script. After some quick googling, I saw some discussion that webscraping with a browser automation tool may be using too powerful of a tool for the desired data collection. At the time, I was interested in poking around in Go, so a quick ‘Go Webscraping’ search later, I found myself point at GoColly. Understand that this webscraping script is my first real Go code I’ve written, so some style may be forgone as I still cling onto my jumbled internalized program representation.
If I’m doing some webscraping, lets peek into what an IS page on openworks looks like
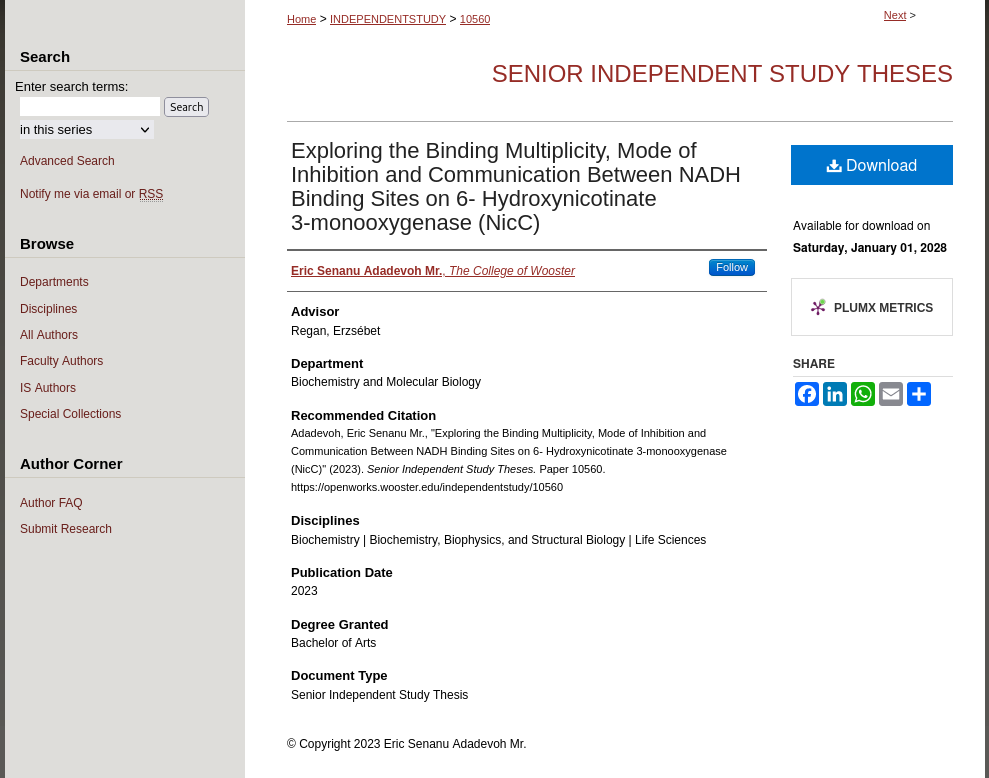
Colly, from the limited usage I interacted with it, feels almost ‘functional’-style in how it operates. Lets do a quick look-over to haskell:
fib1 :: (Integer, Integer) -> Integer
fib1 0 = 1
fib1 1 = 1
fib1 n = fib1(n - 1) * fib2(n - 2)
-- Or, in my very rudimentary haskell knowledge, we can understand pattern matching like so:
<func_name> :: <Types...> -> <ReturnType>
<func_name> <pattern1> = <response1>
<func_name> <pattern2> = <response2>
...
Long story short, we are defining certain patterns that, when encountered, have certain responses. For a fibonacci, if we reach 0 or 1, we are a known base case, otherwise for an unknown value, the response is a recursive functional call. It makes sense… to me… somewhat. Maybe this functional foray helps, perhaps not. But lets look at the main body of the webscraping code:
// define our collector variable
c := colly.NewCollector(
// Don't go OOB
colly.AllowedDomains("openworks.wooster.edu"),
)
// rate limits
c.Limit(&colly.LimitRule{
DomainGlob: "*openworks.wooster.*",
RandomDelay: 5 * time.Second,
})
// debugging purposes
c.OnResponse(func(r *colly.Response) {
fmt.Println("\tVisited", r.Request.URL)
})
// Find and print all links in main body page, visit them
c.OnHTML("p.article-listing", func (e *colly.HTMLElement) {
link := e.ChildAttr("a", "href")
fmt.Println("Visiting: ", link)
c.Visit(link)
})
// pagination
c.OnHTML("div.adjacent-pagination", func (e *colly.HTMLElement) {
currentPage := e.ChildText("li.active a")
fmt.Println("Current page is ", currentPage)
nextpg := nextPageStr(currentPage)
fstr := "div.adjacent-pagination li a[title='" + nextpg + "']"
nextPg := e.ChildAttr(fstr, "href")
c.Visit(nextPg)
})
// parse all data from metadata page
c.OnHTML("div#content", func (e *colly.HTMLElement) {
downl := e.ChildAttr("a[id='pdf']", "href")
titl := e.ChildText("div#title > p")
aut := e.ChildText("p.author > a > strong")
abs := e.ChildText("div#abstract > p")
adv := e.ChildText("div#advisor1 > p")
dept := e.ChildText("div#subject_area > p")
reccite := e.ChildText("div#recommended_citation > p")
keys := e.ChildText("div#keywords > p")
pubdate := e.ChildText("div#publication_date > p")
deg := e.ChildText("div#degree_granted > p")
doctype := e.ChildText("div#document_type > p")
// strip all tabs
titl = replacer.Replace(titl)
aut = replacer.Replace(aut)
abs = replacer.Replace(abs)
adv = replacer.Replace(adv)
dept = replacer.Replace(dept)
reccite = replacer.Replace(reccite)
keys = replacer.Replace(keys)
pubdate = replacer.Replace(pubdate)
deg = replacer.Replace(deg)
doctype = replacer.Replace(doctype)
paper := paperMeta{downl, titl, aut, abs, adv, dept, reccite, keys, pubdate, deg, doctype}
})
c.visit("https://openworks.wooster.edu/independentstudy/")
Notably, we define these various patterns and specific responses that should occur when they are encountered. I.e. c.OnHTML("<Go Selector>", func funcToCall (e *colly.HTMLElement{...})). The last part of this is the concept of ‘Go Selectors’ – which are analagous to CSS Selectors, AFAIK. If you’ve done any web automation, there are many different ways to access various elements on the page (XPaths, Selectors, etc.), so while I am most comfortable with XPaths, trawling some simple Selectors with the aid of developer tools was trivial.
Our various encounterings of elements doesn’t matter which order we define them in, which was the weirdest feeling for me. I really only needed 3 patterns to follow: follow pagination on the listings of IS pages, follow links to individual metadata pages, and harvest the data itself on those metadata pages.
Ok, so if we are webscraping pages that look like the graphic above, lets make a container for an individual IS in underlying Go Lang:
type paperMeta struct {
downlink string
title string
author string
abstract string
advisor string
department string
reccitation string
keywords string
pubdate string
degree string
papertype string
}
alrighty… that isn’t so bad! Structs seem pretty approachable, adjacent to C-style structs and arguably analagous to python’s dataclasses. Throw in some information cleaning (stripping escape and spacing characters, for example), write all of them to file, and I now have ~11,600 observations for normal Independent Studies. If I just swap where the outfile goes, and change where the initial webscraper is pointed at, I got another 375 exemplar independent studies quite easily.
But, this is all metadata. All I have is a TSV datafile, without any of the fun information, like the underlying rawtexts of all these independent studies. To do get those, I needed to somehow download all the papers found at the links in the harvested metadata.
Selenium PDF Farm
No where near as long, nor as interesting of a subproblem to tackle. Given a long list of links to downloadable PDF forms of Independent Studies, I needed to go and download them. The one complication is that observing these rawtexts is behind an authentication step, as many are limited to only Wooster students (a primary reason why this dataset is NOT published for anyone’s free use). My hackish workaround was to just let me manually log in before yielding control to the automation tool for download requests to all the different links on the page. The only semi-unique step here was adding extra profile steps so that pdf files would be automatically downloaded to a pre-specified folder (for me, an external drive):
profile = {"plugins.plugins_list": [{"enabled": False,
"name": "Chrome PDF Viewer"}],
"download.default_directory" : download_folder,
"download.prompt_for_download" : False,
"download.directory_upgrade" : True,
"download.extensions_to_open" : "",
"plugins.always_open_pdf_externally" : True,
}
options.add_experimental_option("prefs", profile)
I now have all the data collected, but not in an easily analyzable form for the stats project, nor in an easily tunable form for an AI, as far as I can tell.
Of note: I was only able to download rawtexts of ~5,500 Independent studies (both exemplar and non), due to embargo rules. There seems to be a blanket limitation for any IS from 2005 or earlier that makes them unable to be accessed.
Lexical Analysis
I snuffed out a textcomplexity python package on pip, but documentation on how to use it was relatively lax. Thankfully, due to the beauty of python, jupyter notebooks, a lonely and minorly alcoholic friday night, and smashing my head against the keyboard for 2 hours, I had a complete analysis script. It wasn’t good, but functional. At a high level, for each PDF, it:
- Parsed the PDF with PyPDF2
- Extracted all text from the document
- Counted all pictures in the document
- Did some metadata cross referencing with invariants on the openworks generated front page
- Handed the raw text into a
StanzaNLP pipeline to convert text to CONLL-U format - Tossed the Conll-U into
textcomplexityto get the end lexical statistics about the IS rawtext - Wrote all data to a file
There are many opportunities for failure here, so my script was littered with try/except control flow handling, which slowed down the script but made it more resilient to failures. So… I started running this on my laptop, and went to bed.
Well.
That.
Sucks?!?!
Further digging into the script, and a desire to be able to use my laptop for like… schoolwork… I transitioned the jupyter notebook over to my Linux Desktop. Creating an entirely new python venv, redownloading all the dependencies… up and running again! So now, I ran the script and got more successes, but each paper was taking 5-10 minutes. Entirely too long! Some further digging, and I quickly identified the largest timesink of the program is converting rawtexts to .conllu format. Inside a stanza script, I wasn’t utilizing my GPU, for dependency parsing, tokenizing, etc. etc.
Ok, GPU acceleration enabled on a GTX 1060 6GB (my computer is old, hush hush). Start this in the background and… run for 72 hours. Ran through all of the ISes at least! But with about a 50 % fail rate, primarily from 50% of the papers unable to be processed and converted to .conllu. I diagnose the error to be CUDA running out of memory. So obviously, the correct solution here is to
A week later, I have an RTX 3060 12 GB swapped in. Lets run this script one more time… and, same issue. CUDA Out Of Memory Error: ... Wants 17 GB (???), 1 GB Available
Hmm. Maybe I should read the error stacktrace. Ah, its coming from Stanza’s POS pipeline segment. Maybe I should look at documentation about how to fix this.
Ok, lets set some limits on the pipeline processor:
nlp = stanza.Pipeline(args.language,
processors="tokenize,pos,lemma,depparse",
download_method=None,
use_gpu=True, # lets use GPU
logging_level='ERROR',
pos_batch_size=1000 # limit part-of-speech batch size
)
Alright. Run this, still running into a few issues. A final workaround was to design a two-tiered processing pattern: First attempt to have the GPU analyze a paper, but if that fails, throw the CPU (and its much larger memory pool) at it compared to the GPU. After running this, I had significantly more luck with the analysis script, with an error rate of 15 some papers, ~<1%, compared to 75%+ in the first pass and 50% in the second pass.
If you are a visual person, here is a graphic the sums up this entire analysis pipeline step:
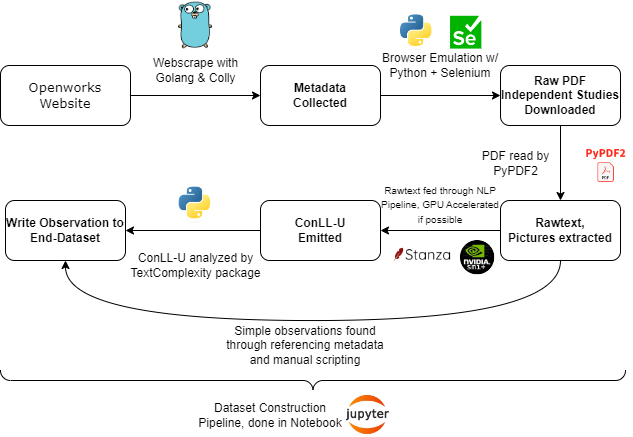
Results
You can read the first draft of the paper.
The final version is complete now too.
Finally, if you are interested in the raw R code used to generate the graphics in the above papers in addition to many graphics that didn’t make it into the final (8 page limited) paper, I am also making the project appendix public.
If you are interested in the dataset or analysis pipeline itself, reach out to me directly. Thanks for reading!